# pip install pandas numpy matplotlib seaborn scikit-learn lifelines scipyimport osimport numpy as npimport pandas as pdimport matplotlib.pyplot as pltimport seaborn as snsfrom sklearn.model_selection import train_test_splitfrom sklearn.preprocessing import StandardScaler, LabelEncoderfrom sklearn.svm import SVC, SVRfrom sklearn.metrics import roc_auc_score, brier_score_lossfrom sklearn.inspection import permutation_importancefrom lifelines import CoxPHFitter, KaplanMeierFitterfrom lifelines.statistics import logrank_testfrom lifelines.calibration import survival_probability_calibrationfrom lifelines.utils import concordance_indeximport scipy.stats as statsfrom scipy.interpolate import interp1dimport warningswarnings.filterwarnings('ignore')# 设置中文字体和路径plt.rcParams['font.sans-serif'] = ['SimHei', 'Arial Unicode MS']plt.rcParams['axes.unicode_minus'] = False# 创建结果目录results_dir = os.path.expanduser("~/Desktop/Results模型-svm-poly")os.makedirs(results_dir, exist_ok=True)class PolynomialSVM_Survival: def __init__(self, degree=2, C=1.0, kernel='poly'): self.degree = degree self.C = C self.kernel = kernel self.scaler = StandardScaler() self.model = None self.model_type = None def prepare_data(self, data, features, time_col='时间', event_col='结局'):"""准备数据"""# 选择特征 X = data[features].copy() self.feature_names = features# 处理分类变量for col in X.columns:if X[col].dtype == 'object': le = LabelEncoder() X[col] = le.fit_transform(X[col].astype(str))# 处理缺失值 X = X.fillna(X.median())# 获取生存数据 T = data[time_col].values E = data[event_col].valuesreturn X, T, E def fit(self, data, features, time_col='时间', event_col='结局'):"""训练模型""" X, T, E = self.prepare_data(data, features, time_col, event_col)# 标准化特征 X_scaled = self.scaler.fit_transform(X)# 尝试不同的SVM方法 try:

return self def predict_risk(self, data, features):"""预测风险评分""" X, _, _ = self.prepare_data(data, features) X_scaled = self.scaler.transform(X)if self.model_type == "多项式核SVR回归":# 对于回归,风险评分与预测时间成反比 predictions = self.model.predict(X_scaled) risk_scores = -predictions # 时间越短风险越高else:# 对于分类,使用决策函数或概率if hasattr(self.model, 'decision_function'): risk_scores = self.model.decision_function(X_scaled)else: risk_scores = self.model.predict_proba(X_scaled)[:, 1]return risk_scores def evaluate(self, train_data, test_data, features, time_col='时间', event_col='结局'):"""评估模型性能"""# 预测风险评分 train_risk = self.predict_risk(train_data, features) test_risk = self.predict_risk(test_data, features)# 获取生存数据 _, train_T, train_E = self.prepare_data(train_data, features, time_col, event_col) _, test_T, test_E = self.prepare_data(test_data, features, time_col, event_col)# 计算C-index train_cindex = concordance_index(train_T, -train_risk, train_E) test_cindex = concordance_index(test_T, -test_risk, test_E)print(f"训练集C-index: {train_cindex:.3f}")print(f"测试集C-index: {test_cindex:.3f}")return {'train_risk': train_risk,'test_risk': test_risk,'train_cindex': train_cindex,'test_cindex': test_cindex,'train_T': train_T,'train_E': train_E,'test_T': test_T,'test_E': test_E }def time_dependent_auc(T, E, risk_scores, time_points):"""计算时间依赖性AUC""" auc_results = {}for time_point in time_points: try:# 创建该时间点的标签# 事件发生在时间点之前为1,删失或事件发生在之后为0 y_true = ((T <= time_point) & (E == 1)).astype(int)# 只考虑在时间点之前有风险的患者 at_risk = (T >= time_point) | ((T <= time_point) & (E == 1))if len(np.unique(y_true[at_risk])) > 1: auc = roc_auc_score(y_true[at_risk], risk_scores[at_risk]) auc_results[time_point] = aucprint(f"{time_point}个月AUC: {auc:.3f}")else: auc_results[time_point] = 0.5print(f"{time_point}个月AUC: 0.500 (无法计算)") except Exception as e:print(f"时间点{time_point}个月AUC计算失败: {e}") auc_results[time_point] = np.nanreturn auc_resultsdef calculate_brier_score(T, E, risk_scores, time_points):"""计算Brier分数""" brier_scores = {}for time_point in time_points: try:# 创建观测结果 observed = ((T <= time_point) & (E == 1)).astype(int) at_risk = (T >= time_point) | ((T <= time_point) & (E == 1))# 将风险评分转换为生存概率 risk_norm = (risk_scores - np.min(risk_scores)) / (np.max(risk_scores) - np.min(risk_scores)) predicted_survival = 1 - risk_normif len(observed[at_risk]) > 0: brier_score = brier_score_loss(observed[at_risk], predicted_survival[at_risk]) brier_scores[time_point] = brier_scoreprint(f"{time_point}个月Brier分数: {brier_score:.3f}")else: brier_scores[time_point] = np.nan except Exception as e:print(f"时间点{time_point}个月Brier分数计算失败: {e}") brier_scores[time_point] = np.nanreturn brier_scoresdef create_visualizations(eval_results, features, test_data, results_dir):"""创建所有可视化图表"""
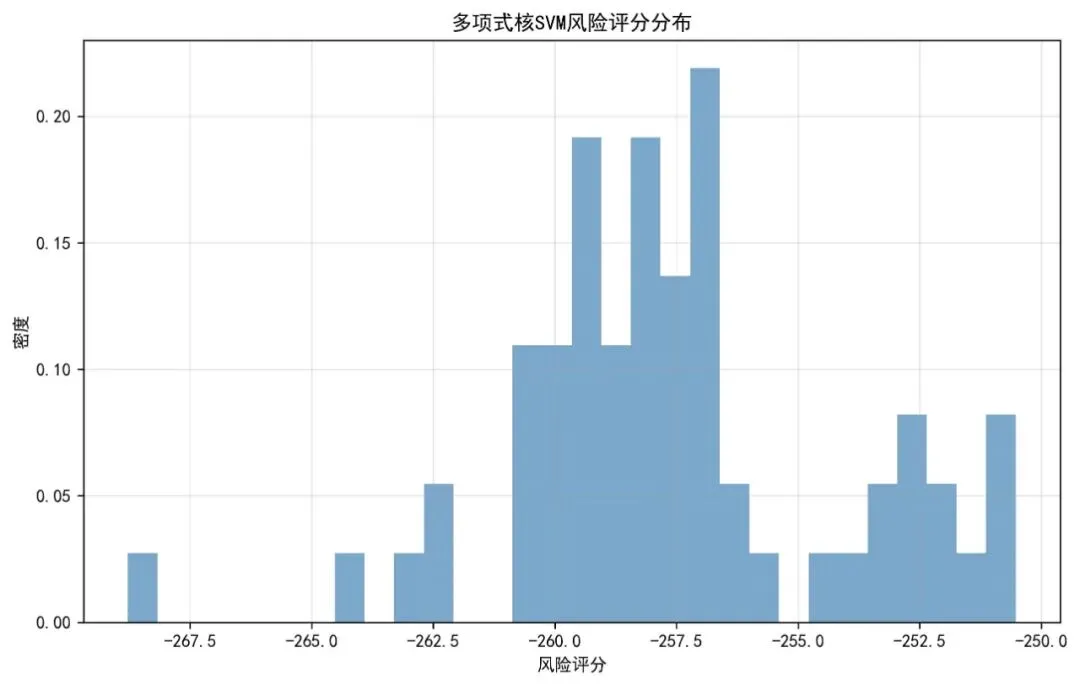
# 1. 风险评分分布图 plt.figure(figsize=(10, 6)) plt.hist(eval_results['test_risk'], bins=30, density=True, alpha=0.7, color='steelblue') plt.xlabel('风险评分') plt.ylabel('密度') plt.title('多项式核SVM风险评分分布') plt.grid(True, alpha=0.3) plt.savefig(os.path.join(results_dir, '风险评分分布图.jpg'), dpi=300, bbox_inches='tight') plt.close()
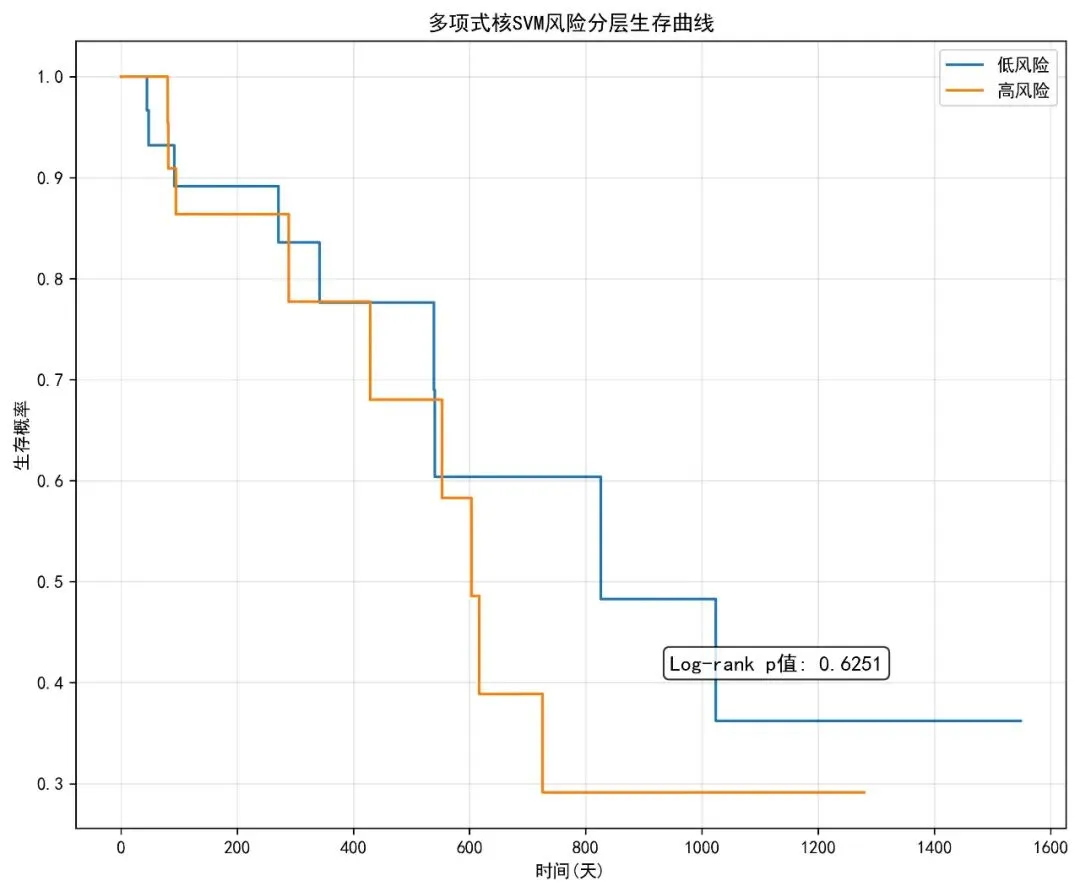
# 2. 风险分层生存曲线 test_risk = eval_results['test_risk'] risk_groups = ['低风险'if x <= np.median(test_risk) else'高风险'for x in test_risk] plt.figure(figsize=(10, 8)) kmf = KaplanMeierFitter()for group in ['低风险', '高风险']: mask = np.array(risk_groups) == group kmf.fit(eval_results['test_T'][mask], eval_results['test_E'][mask], label=group) kmf.plot(ci_show=False)# 添加log-rank检验p值 low_risk_mask = np.array(risk_groups) == '低风险' results = logrank_test(eval_results['test_T'][low_risk_mask], eval_results['test_T'][~low_risk_mask], eval_results['test_E'][low_risk_mask], eval_results['test_E'][~low_risk_mask]) plt.text(0.6, 0.2, f'Log-rank p值: {results.p_value:.4f}', transform=plt.gca().transAxes, fontsize=12, bbox=dict(boxstyle="round,pad=0.3", facecolor="white", alpha=0.8)) plt.xlabel('时间(天)') plt.ylabel('生存概率') plt.title('多项式核SVM风险分层生存曲线') plt.grid(True, alpha=0.3) plt.legend() plt.savefig(os.path.join(results_dir, '风险分层生存曲线.jpg'), dpi=300, bbox_inches='tight') plt.close()
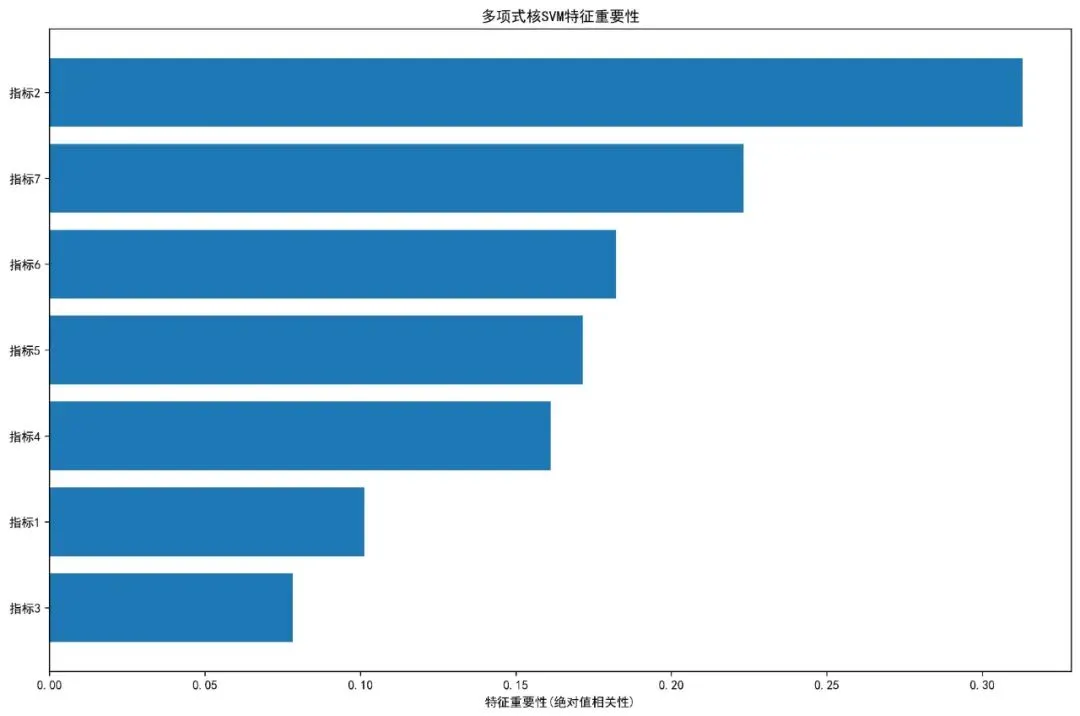
# 3. 特征重要性图if len(features) > 0: plt.figure(figsize=(12, 8))# 使用简单的相关性分析作为特征重要性 feature_importance = []for feature in features:if test_data[feature].dtype in ['int64', 'float64']: corr = np.corrcoef(test_data[feature].fillna(test_data[feature].median()), eval_results['test_risk'])[0, 1] feature_importance.append(abs(corr))else: feature_importance.append(0) importance_df = pd.DataFrame({'Feature': features,'Importance': feature_importance }).sort_values('Importance', ascending=True) plt.barh(importance_df['Feature'], importance_df['Importance']) plt.xlabel('特征重要性(绝对值相关性)') plt.title('多项式核SVM特征重要性') plt.tight_layout() plt.savefig(os.path.join(results_dir, '特征重要性图.jpg'), dpi=300, bbox_inches='tight') plt.close()def generate_report(eval_results, model, time_points, auc_results, brier_results, results_dir):"""生成综合报告"""# 性能指标汇总 performance_df = pd.DataFrame({'指标': ['训练集C-index', '测试集C-index'] + [f'{t}个月AUC'for t in time_points] + [f'{t}个月Brier分数'for tin time_points],'值': [eval_results['train_cindex'], eval_results['test_cindex']] + [auc_results.get(t, np.nan) for t in time_points] + [brier_results.get(t, np.nan) for t in time_points] }) performance_df.to_csv(os.path.join(results_dir, '性能指标.csv'), index=False, encoding='utf-8-sig')# 模型信息 model_info = pd.DataFrame({'参数': ['模型类型', '核函数', '度数', 'C参数', '训练样本数', '测试样本数'],'值': [model.model_type, model.kernel, model.degree, model.C, len(eval_results['train_T']), len(eval_results['test_T'])] }) model_info.to_csv(os.path.join(results_dir, '模型信息.csv'), index=False, encoding='utf-8-sig')# 生成文本报告 with open(os.path.join(results_dir, '分析报告.txt'), 'w', encoding='utf-8') as f: f.write("多项式核SVM生存模型分析报告\n") f.write("=" * 50 + "\n\n") f.write(f"模型类型: {model.model_type}\n") f.write(f"核函数: {model.kernel}\n") f.write(f"度数: {model.degree}\n") f.write(f"C参数: {model.C}\n\n") f.write("性能指标:\n") f.write(f"训练集C-index: {eval_results['train_cindex']:.3f}\n") f.write(f"测试集C-index: {eval_results['test_cindex']:.3f}\n")for t in time_points: f.write(f"{t}个月AUC: {auc_results.get(t, 'N/A')}\n") f.write(f"{t}个月Brier分数: {brier_results.get(t, 'N/A')}\n") f.write("\n关键结论:\n")if eval_results['test_cindex'] > 0.7: f.write("模型表现出优秀的预测能力\n")elif eval_results['test_cindex'] > 0.6: f.write("模型表现出良好的预测能力\n")else: f.write("模型预测能力有待改进\n") f.write("\n模型特点:\n") f.write("- 使用多项式核函数,能够捕捉变量间的非线性关系\n") f.write("- 通过核技巧将数据映射到高维空间\n") f.write("- 适用于复杂的数据模式\n")def main():"""主函数"""print("开始多项式核SVM生存分析...")# 1. 读取数据 try:# 请根据实际数据路径修改 data_path = "示例数据.xlsx"# 修改为实际路径 data = pd.read_excel(data_path, sheet_name="示例数据")print(f"数据读取成功,维度: {data.shape}") except Exception as e:print(f"数据读取失败: {e}")# 创建示例数据用于演示print("创建示例数据用于演示...") np.random.seed(42) n_samples = 200 data = pd.DataFrame({'时间': np.random.exponential(365, n_samples),'结局': np.random.choice([0, 1], n_samples, p=[0.7, 0.3]),'指标1': np.random.normal(50, 10, n_samples),'指标2': np.random.normal(100, 20, n_samples),'指标3': np.random.normal(25, 5, n_samples),'指标4': np.random.normal(75, 15, n_samples),'指标5': np.random.normal(30, 8, n_samples),'指标6': np.random.choice(['A', 'B', 'C'], n_samples),'指标7': np.random.normal(60, 12, n_samples) })# 2. 数据预处理print("数据预处理...")# 处理分类变量 categorical_cols = data.select_dtypes(include=['object']).columnsfor col in categorical_cols: le = LabelEncoder() data[col] = le.fit_transform(data[col].astype(str))# 确保数值类型 data['时间'] = pd.to_numeric(data['时间'], errors='coerce') data['结局'] = pd.to_numeric(data['结局'], errors='coerce')# 移除缺失值 data = data.dropna()print(f"处理后数据维度: {data.shape}")# 3. 选择特征 feature_candidates = ['指标1', '指标2', '指标3', '指标4', '指标5', '指标6', '指标7'] available_features = [f for f in feature_candidates if f in data.columns]print(f"可用特征: {available_features}")# 4. 划分训练集和测试集 train_data, test_data = train_test_split(data, test_size=0.3, random_state=2025, stratify=data['结局'])print(f"训练集样本数: {len(train_data)}")print(f"测试集样本数: {len(test_data)}")# 5. 训练多项式核SVM模型print("训练多项式核SVM模型...") svm_model = PolynomialSVM_Survival(degree=2, C=1.0, kernel='poly') try: svm_model.fit(train_data, available_features, time_col='时间', event_col='结局')# 6. 模型评估print("模型评估...") eval_results = svm_model.evaluate(train_data, test_data, available_features)# 7. 时间依赖性评估 time_points = [12, 24, 36] # 1年、2年、3年print("计算时间依赖性AUC...") auc_results = time_dependent_auc(eval_results['test_T'], eval_results['test_E'], eval_results['test_risk'], time_points)print("计算Brier分数...") brier_results = calculate_brier_score(eval_results['test_T'], eval_results['test_E'], eval_results['test_risk'], time_points)# 8. 可视化print("生成可视化图表...") create_visualizations(eval_results, available_features, test_data, results_dir)# 9. 生成报告print("生成分析报告...") generate_report(eval_results, svm_model, time_points, auc_results, brier_results, results_dir)# 10. 保存风险评分 test_data_with_risk = test_data.copy() test_data_with_risk['风险评分'] = eval_results['test_risk'] test_data_with_risk.to_csv(os.path.join(results_dir, '测试集风险评分.csv'), index=False, encoding='utf-8-sig')print(f"\n=== 多项式核SVM生存分析完成 ===")print(f"结果保存至: {results_dir}")print(f"模型类型: {svm_model.model_type}")print(f"测试集C-index: {eval_results['test_cindex']:.3f}")if eval_results['test_cindex'] < 0.5:print("\n!!! 警告: 测试集C-index低于0.5,模型表现不佳 !!!")print("建议:")print("- 调整SVM参数(度数、C参数等)")print("- 检查数据质量和特征选择")print("- 尝试其他核函数") except Exception as e:print(f"模型训练失败: {e}")print("请检查数据格式和特征选择")if __name__ == "__main__": main()