import osimport pandas as pdimport numpy as npfrom datetime import datetime, timedeltaimport matplotlib.pyplot as pltimport seaborn as snsfrom statsmodels.tsa.statespace.sarimax import SARIMAXfrom statsmodels.tsa.seasonal import seasonal_decomposefrom statsmodels.tsa.stattools import adfullerfrom sklearn.metrics import mean_absolute_error, mean_squared_errorimport warningswarnings.filterwarnings('ignore')def get_desktop_path():"""获取桌面路径"""return os.path.join(os.path.expanduser("~"), "Desktop")def create_results_folder():"""创建结果文件夹""" desktop_path = get_desktop_path() results_path = os.path.join(desktop_path, "Results时间SARIMA")# 创建主文件夹和子文件夹 folders = [ results_path, os.path.join(results_path, "Models"), os.path.join(results_path, "Predictions"), os.path.join(results_path, "Diagnostics"), os.path.join(results_path, "Seasonal_Plots"), os.path.join(results_path, "Summary_Reports") ]for folder in folders:if not os.path.exists(folder): os.makedirs(folder)print(f"创建文件夹: {folder}")return results_pathdef load_and_prepare_data(results_path):"""加载和准备数据"""print("正在加载数据...")# 尝试读取R生成的数据 try: data_path = os.path.join(get_desktop_path(), "Results", "combined_weather_disease_data.csv") combined_data = pd.read_csv(data_path) combined_data['timestamp'] = pd.to_datetime(combined_data['timestamp'])print(f"成功读取数据,共{len(combined_data)}行")return combined_data except FileNotFoundError:print("未找到R生成的数据文件,生成模拟数据...")return generate_sample_data()def generate_sample_data():"""生成示例数据(如果R数据不存在)""" dates = pd.date_range('1981-01-01', '2025-12-31', freq='D')# 生成模拟疾病数据 np.random.seed(42) n = len(dates) data = {'timestamp': dates,'year': dates.year,'month': dates.month,'day': dates.day, }# 生成季节性疾病数据 t = np.arange(n)# 流感 - 冬季高发 data['influenza'] = np.round( 50 + 30 * np.sin(2 * np.pi * (t - 10) / 365) + # 冬季高峰 10 * np.sin(2 * np.pi * (t - 200) / 365) + # 夏季小高峰 np.random.normal(0, 5, n) ).clip(0)# 手足口病 - 春夏季高发 data['hand_foot_mouth'] = np.round( 30 + 20 * np.sin(2 * np.pi * (t - 150) / 365) + # 春夏季高峰 8 * np.sin(2 * np.pi * (t - 330) / 365) + # 秋季小高峰 np.random.normal(0, 3, n) ).clip(0)# 细菌性痢疾 - 夏季高发 data['bacillary_dysentery'] = np.round( 25 + 15 * np.sin(2 * np.pi * (t - 180) / 365) + # 夏季高峰 np.random.normal(0, 4, n) ).clip(0)# 流行性出血热 - 春秋季高发 data['hemorrhagic_fever'] = np.round( 20 + 12 * np.sin(2 * np.pi * (t - 80) / 365) + # 春季高峰 10 * np.sin(2 * np.pi * (t - 280) / 365) + # 秋季高峰 np.random.normal(0, 2, n) ).clip(0)return pd.DataFrame(data)def prepare_monthly_data(daily_data, disease_columns):"""将日数据转换为月数据""" monthly_data = daily_data.set_index('timestamp').resample('M')[disease_columns].sum().reset_index() monthly_data['year'] = monthly_data['timestamp'].dt.year monthly_data['month'] = monthly_data['timestamp'].dt.monthreturn monthly_data
def forecast_and_evaluate(model, test_data, disease_name):"""进行预测和评估"""if model is None:return None# 预测 forecast_steps = len(test_data) forecast_result = model.get_forecast(steps=forecast_steps) forecast_mean = forecast_result.predicted_mean confidence_intervals = forecast_result.conf_int()# 计算评估指标 actual = test_data[disease_name].values predicted = forecast_mean.values mae = mean_absolute_error(actual, predicted) rmse = np.sqrt(mean_squared_error(actual, predicted)) mape = np.mean(np.abs((actual - predicted) / actual)) * 100 results = {'actual': actual,'predicted': predicted,'lower_95': confidence_intervals.iloc[:, 0].values,'upper_95': confidence_intervals.iloc[:, 1].values,'mae': mae,'rmse': rmse,'mape': mape }return resultsdef create_forecast_plot(monthly_data, train_data, test_data, results, disease_name, results_path):"""创建预测图""" plt.figure(figsize=(14, 8))# 完整时间序列 full_dates = monthly_data['timestamp'] full_values = monthly_data[disease_name]# 训练数据 train_mask = full_dates < pd.Timestamp('2016-01-01') plt.plot(full_dates[train_mask], full_values[train_mask], color='blue', linewidth=1, label='训练数据')# 测试数据(实际值) test_mask = full_dates >= pd.Timestamp('2016-01-01') plt.plot(full_dates[test_mask], full_values[test_mask], color='darkgreen', linewidth=1, label='实际值')# 预测值 forecast_dates = test_data['timestamp'] plt.plot(forecast_dates, results['predicted'], color='red', linewidth=1.5, label='预测值')# 预测区间 plt.fill_between(forecast_dates, results['lower_95'], results['upper_95'], color='red', alpha=0.2, label='95%置信区间')# 分割线 plt.axvline(x=pd.Timestamp('2016-01-01'), color='gray', linestyle='--', alpha=0.7) plt.title(f'{disease_name} SARIMA模型预测结果', fontsize=16, fontweight='bold') plt.xlabel('日期', fontsize=12) plt.ylabel('病例数', fontsize=12) plt.legend() plt.grid(True, alpha=0.3) plt.xticks(rotation=45)# 保存图片 plot_path = os.path.join(results_path, "Predictions", f"SARIMA_Forecast_{disease_name}.png") plt.tight_layout() plt.savefig(plot_path, dpi=300, bbox_inches='tight') plt.close()print(f"预测图已保存: {plot_path}")
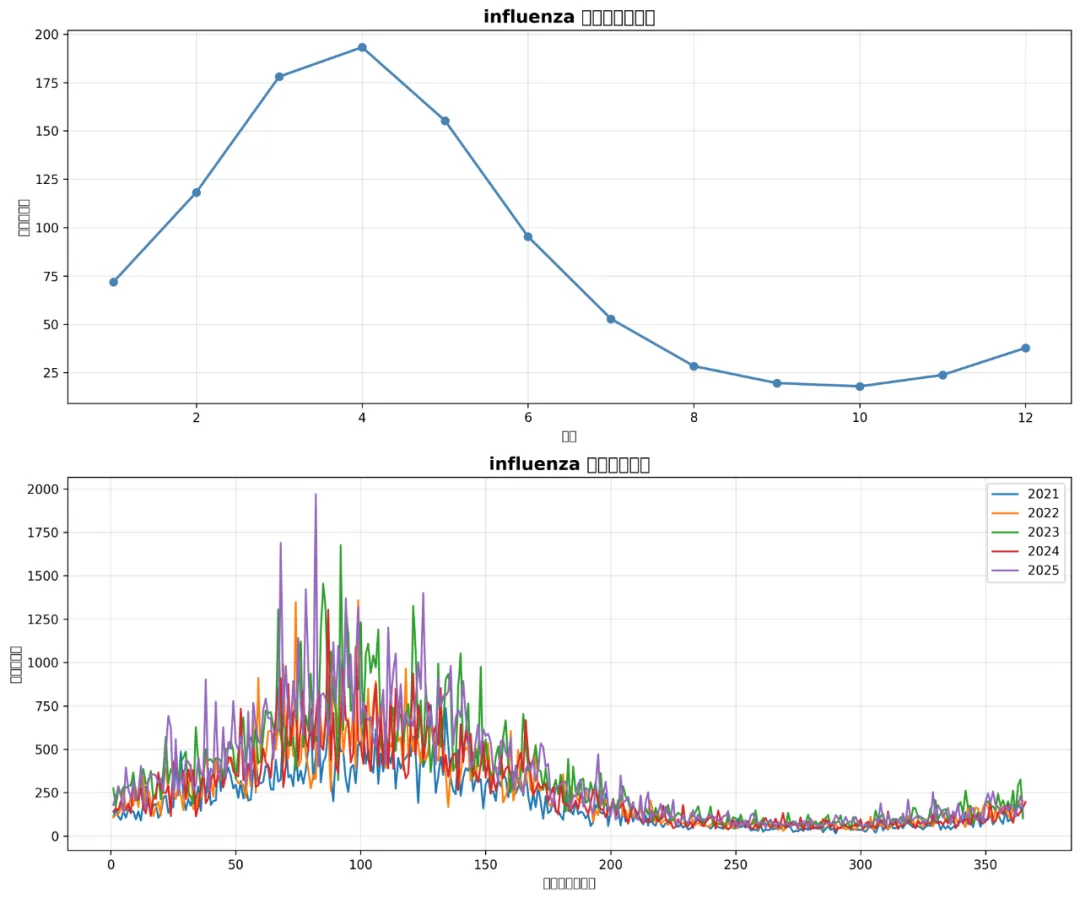
def create_seasonal_plots(daily_data, disease_name, results_path):"""创建季节性分析图"""# 月度季节性模式 monthly_seasonal = daily_data.groupby('month')[disease_name].mean().reset_index()# 年度对比图(最近5年) recent_years = range(daily_data['year'].max() - 4, daily_data['year'].max() + 1) yearly_data = daily_data[daily_data['year'].isin(recent_years)].copy() yearly_data['day_of_year'] = yearly_data['timestamp'].dt.dayofyear yearly_avg = yearly_data.groupby(['year', 'day_of_year'])[disease_name].mean().reset_index()# 创建子图 fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 10))# 月度季节性图 ax1.plot(monthly_seasonal['month'], monthly_seasonal[disease_name], marker='o', linewidth=2, color='steelblue') ax1.set_title(f'{disease_name} 月度季节性模式', fontsize=14, fontweight='bold') ax1.set_xlabel('月份') ax1.set_ylabel('平均病例数') ax1.grid(True, alpha=0.3)# 年度对比图for year in recent_years: year_data = yearly_avg[yearly_avg['year'] == year] ax2.plot(year_data['day_of_year'], year_data[disease_name], label=str(year), linewidth=1.5) ax2.set_title(f'{disease_name} 年度模式对比', fontsize=14, fontweight='bold') ax2.set_xlabel('一年中的第几天') ax2.set_ylabel('平均病例数') ax2.legend() ax2.grid(True, alpha=0.3) plt.tight_layout()# 保存图片 plot_path = os.path.join(results_path, "Seasonal_Plots", f"Seasonal_Analysis_{disease_name}.png") plt.savefig(plot_path, dpi=300, bbox_inches='tight') plt.close()print(f"季节性分析图已保存: {plot_path}")
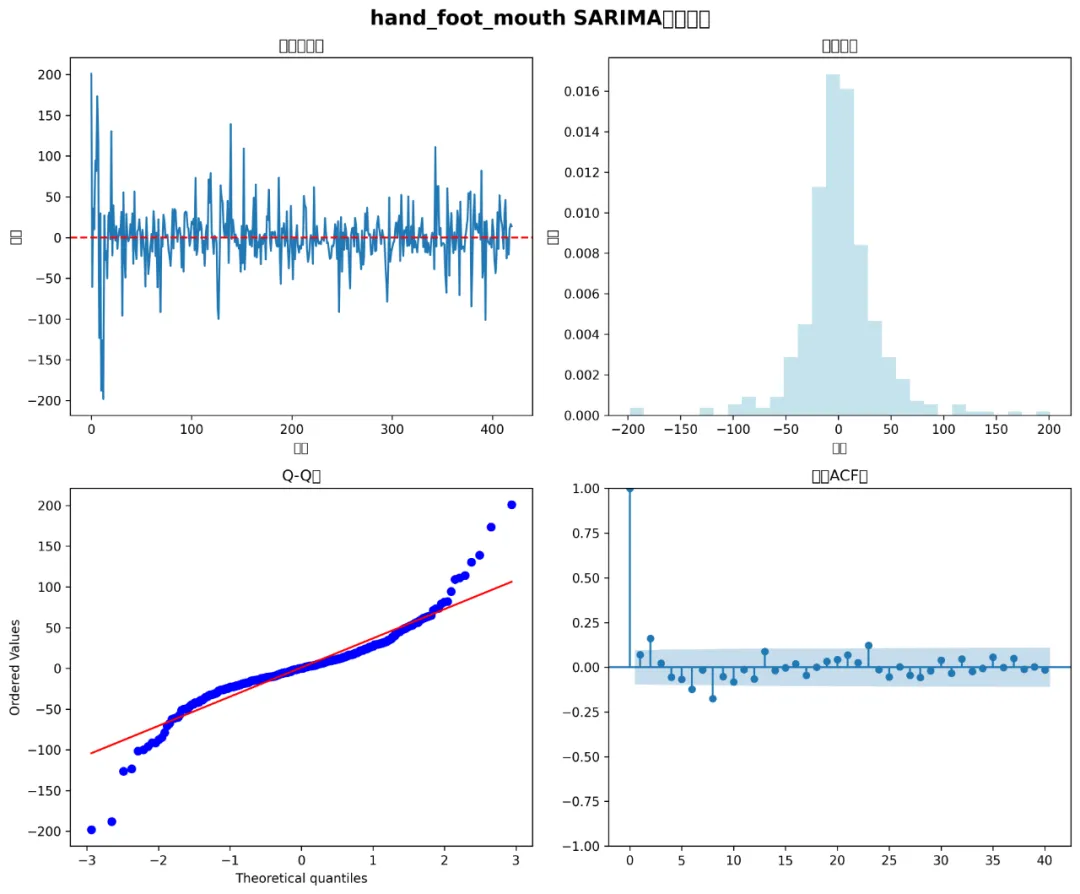
def create_diagnostic_plots(model, disease_name, results_path):"""创建模型诊断图"""if model is None:return# 残差分析 residuals = model.resid fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2, figsize=(12, 10))# 残差时序图 ax1.plot(residuals) ax1.set_title('残差时序图') ax1.set_xlabel('时间') ax1.set_ylabel('残差') ax1.axhline(y=0, color='r', linestyle='--')# 残差直方图 ax2.hist(residuals, bins=30, density=True, alpha=0.7, color='lightblue') ax2.set_title('残差分布') ax2.set_xlabel('残差') ax2.set_ylabel('密度')# Q-Q图 from scipy import stats stats.probplot(residuals, dist="norm", plot=ax3) ax3.set_title('Q-Q图')# ACF图 from statsmodels.graphics.tsaplots import plot_acf plot_acf(residuals, ax=ax4, lags=40) ax4.set_title('残差ACF图') plt.suptitle(f'{disease_name} SARIMA模型诊断', fontsize=16, fontweight='bold') plt.tight_layout()# 保存图片 plot_path = os.path.join(results_path, "Diagnostics", f"SARIMA_Diagnostics_{disease_name}.png") plt.savefig(plot_path, dpi=300, bbox_inches='tight') plt.close()print(f"诊断图已保存: {plot_path}")def main():"""主函数"""print("开始SARIMA建模分析...")# 创建结果文件夹 results_path = create_results_folder()print(f"结果将保存到: {results_path}")# 加载数据 combined_data = load_and_prepare_data(results_path)# 选择分析的疾病 selected_diseases = ["influenza", # 流感"hand_foot_mouth", # 手足口病"bacillary_dysentery", # 细菌性痢疾"hemorrhagic_fever"# 流行性出血热 ]# 准备月度数据 monthly_data = prepare_monthly_data(combined_data, selected_diseases)# 划分训练集和测试集 train_data = monthly_data[monthly_data['timestamp'] < '2016-01-01'] test_data = monthly_data[monthly_data['timestamp'] >= '2016-01-01']print(f"训练集: {len(train_data)} 个月 (1981-2015)")print(f"测试集: {len(test_data)} 个月 (2016-2025)")# 存储结果 all_results = {} accuracy_summary = []# 为每种疾病拟合模型for disease in selected_diseases:print(f"\n{'=' * 50}")print(f"处理疾病: {disease}")print(f"{'=' * 50}")# 拟合SARIMA模型 model = fit_sarima_model(train_data[disease], disease)if model is not None:# 预测和评估 results = forecast_and_evaluate(model, test_data, disease)if results is not None: all_results[disease] = {'model': model,'results': results }# 创建可视化 create_forecast_plot(monthly_data, train_data, test_data, results, disease, results_path) create_seasonal_plots(combined_data, disease, results_path) create_diagnostic_plots(model, disease, results_path)# 保存精度结果 accuracy_summary.append({'Disease': disease,'MAE': round(results['mae'], 2),'RMSE': round(results['rmse'], 2),'MAPE': round(results['mape'], 2) })# 保存预测结果 forecast_df = pd.DataFrame({'date': test_data['timestamp'],'actual': results['actual'],'predicted': results['predicted'],'lower_95': results['lower_95'],'upper_95': results['upper_95'] }) forecast_path = os.path.join(results_path, "Predictions", f"SARIMA_Predictions_{disease}.csv") forecast_df.to_csv(forecast_path, index=False)print(f"预测结果已保存: {forecast_path}")# 生成汇总报告if accuracy_summary:# 精度汇总 accuracy_df = pd.DataFrame(accuracy_summary) accuracy_path = os.path.join(results_path, "Summary_Reports", "accuracy_summary.csv") accuracy_df.to_csv(accuracy_path, index=False)# 创建精度比较图 plt.figure(figsize=(10, 6)) bars = plt.bar(accuracy_df['Disease'], accuracy_df['MAPE'], alpha=0.7) plt.title('SARIMA模型预测精度比较 (MAPE%)', fontsize=14, fontweight='bold') plt.xlabel('疾病类型') plt.ylabel('平均绝对百分比误差 (%)') plt.xticks(rotation=45)# 在柱子上添加数值for bar, mape in zip(bars, accuracy_df['MAPE']): plt.text(bar.get_x() + bar.get_width() / 2, bar.get_height() + 0.5, f'{mape:.1f}%', ha='center', va='bottom') plt.tight_layout() accuracy_plot_path = os.path.join(results_path, "Summary_Reports", "accuracy_comparison.png") plt.savefig(accuracy_plot_path, dpi=300, bbox_inches='tight') plt.close()print(f"\n精度汇总已保存: {accuracy_path}")print(f"精度比较图已保存: {accuracy_plot_path}")# 显示汇总结果print("\n" + "=" * 60)print("SARIMA建模分析完成!")print("=" * 60)print(f"成功分析的疾病: {list(all_results.keys())}")print(f"\n模型性能摘要:")print(accuracy_df.to_string(index=False))else:print("\n警告: 没有成功拟合任何模型!")print(f"\n所有结果已保存到: {results_path}")if __name__ == "__main__": main()