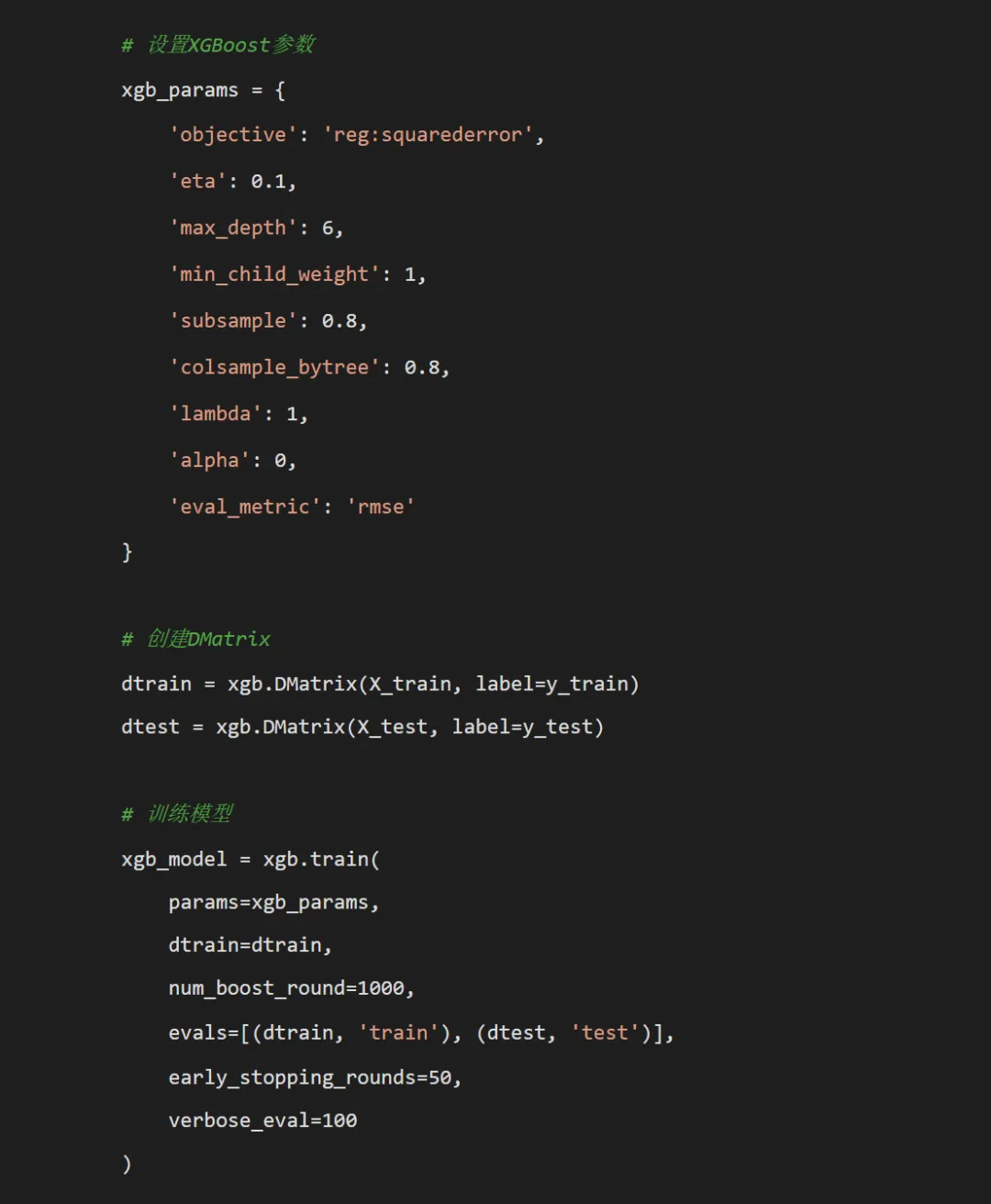
import osimport pandas as pdimport numpy as npimport xgboost as xgbfrom sklearn.metrics import mean_squared_error, mean_absolute_error, r2_scorefrom sklearn.model_selection import train_test_splitimport matplotlib.pyplot as pltimport seaborn as snsimport shapfrom datetime import datetimeimport warningswarnings.filterwarnings('ignore')# 设置中文字体plt.rcParams['font.sans-serif'] = ['SimHei', 'Microsoft YaHei', 'DejaVu Sans']plt.rcParams['axes.unicode_minus'] = Falsedef main():# 获取桌面路径 desktop_path = os.path.join(os.path.expanduser("~"), "Desktop") data_path = os.path.join(desktop_path, "Results时间", "combined_weather_disease_data.csv") results_path = os.path.join(desktop_path, "Results时间XGB")print("开始XGBoost疾病预测建模...")# 检查数据文件是否存在if not os.path.exists(data_path):print(f"错误: 数据文件不存在于 {data_path}")print("请确保桌面上的 'Results时间' 文件夹中包含 'combined_weather_disease_data.csv' 文件")return# 创建子文件夹 subfolders = ["Models", "Predictions", "Plots", "Feature_Importance","Partial_Dependence", "SHAP_Analysis"]for folder in subfolders: folder_path = os.path.join(results_path, folder)if not os.path.exists(folder_path): os.makedirs(folder_path)# 读取数据 try:print(f"从 {data_path} 读取数据...") combined_data = pd.read_csv(data_path) combined_data['timestamp'] = pd.to_datetime(combined_data['timestamp'])print("数据读取成功!")print(f"数据形状: {combined_data.shape}")print(f"数据列名: {list(combined_data.columns)}") except Exception as e:print(f"读取数据时出错: {e}")return# 选择要建模的疾病 target_diseases = ["influenza", "common_cold", "pneumonia","bacillary_dysentery", "hand_foot_mouth","hemorrhagic_fever"]# 检查目标疾病列是否存在 available_diseases = [disease for disease in target_diseases if disease in combined_data.columns]if not available_diseases:print("错误: 数据中未找到任何目标疾病列")print(f"数据中的列: {list(combined_data.columns)}")returnprint("目标疾病:", ", ".join(available_diseases)) def create_features(data):"""特征工程函数""" df = data.copy()# 时间特征 df['year'] = df['timestamp'].dt.year df['month'] = df['timestamp'].dt.month df['day'] = df['timestamp'].dt.day df['week'] = df['timestamp'].dt.isocalendar().week df['day_of_week'] = df['timestamp'].dt.dayofweek + 1 # 1-7, 1=周一 df['quarter'] = df['timestamp'].dt.quarter df['day_of_year'] = df['timestamp'].dt.dayofyear df['is_weekend'] = (df['day_of_week'].isin([6, 7])).astype(int)# 季节特征 conditions = [ df['month'].isin([12, 1, 2]), df['month'].isin([3, 4, 5]), df['month'].isin([6, 7, 8]), df['month'].isin([9, 10, 11]) ] choices = ['Winter', 'Spring', 'Summer', 'Fall'] df['season'] = np.select(conditions, choices, default='Unknown')# 滞后特征 lag_periods = [1, 7, 30] weather_cols = ['temp_mean', 'humidity_mean', 'precipitation_total', 'sunshine_hours']for col in weather_cols:if col in df.columns:for lag in lag_periods:if lag <= 30: # 避免过多的NA df[f'{col}_lag{lag}'] = df[col].shift(lag)# 滚动窗口特征 window_sizes = [7, 30]for col in ['temp_mean', 'humidity_mean']:if col in df.columns:for window in window_sizes: df[f'{col}_rollmean{window}'] = df[col].rolling(window=window, min_periods=1).mean()for window in window_sizes:if'precipitation_total'in df.columns: df[f'precipitation_rollsum{window}'] = df['precipitation_total'].rolling(window=window, min_periods=1).sum()# 季节性特征 df['sin_day'] = np.sin(2 * np.pi * df['day_of_year'] / 365) df['cos_day'] = np.cos(2 * np.pi * df['day_of_year'] / 365) df['sin_week'] = np.sin(2 * np.pi * df['week'] / 52) df['cos_week'] = np.cos(2 * np.pi * df['week'] / 52)# 为每种疾病创建滞后特征for disease in available_diseases:if disease in df.columns:for lag in [1, 7, 30]: df[f'{disease}_lag{lag}'] = df[disease].shift(lag)for window in [7, 30]: df[f'{disease}_rollmean{window}'] = df[disease].rolling(window=window, min_periods=1).mean()# 移除由于滞后产生的NA df = df.dropna()return df# 应用特征工程print("正在进行特征工程...") featured_data = create_features(combined_data)print(f"特征工程后数据形状: {featured_data.shape}")# 定义基础特征 base_features = [# 气象特征"temp_max", "temp_min", "temp_mean", "temp_median","humidity_max", "humidity_min", "humidity_mean", "humidity_median","wind_max", "wind_mean","pressure_max", "pressure_min", "pressure_mean","precipitation_total", "sunshine_hours",# 时间特征"year", "month", "day", "week", "day_of_week", "day_of_year","is_weekend", "quarter",# 气象滞后特征"temp_mean_lag1", "temp_mean_lag7", "temp_mean_lag30","humidity_mean_lag1", "humidity_mean_lag7", "humidity_mean_lag30","precipitation_lag1", "precipitation_lag7","sunshine_lag1", "sunshine_lag7",# 滑动窗口特征"temp_mean_rollmean7", "temp_mean_rollmean30","humidity_mean_rollmean7", "humidity_mean_rollmean30","precipitation_rollsum7", "precipitation_rollsum30",# 季节性特征"sin_day", "cos_day", "sin_week", "cos_week" ]# 只保留实际存在的特征 base_features = [f for f in base_features if f in featured_data.columns]print(f"可用基础特征数量: {len(base_features)}")# 存储结果 results_summary = [] predictions_list = {} models_list = {} feature_importance_list = {} X_train_list = {}# 对每种疾病进行建模for disease in available_diseases:if disease not in featured_data.columns:print(f"跳过疾病 {disease},数据中不存在该列")continueprint(f"\n正在处理疾病: {disease}")# 添加该疾病的滞后特征 disease_specific_features = base_features + [ f"{disease}_lag1", f"{disease}_lag7", f"{disease}_lag30", f"{disease}_rollmean7", f"{disease}_rollmean30" ]# 只保留存在的特征 available_features = [f for f in disease_specific_features if f in featured_data.columns] available_features.append(disease)# 创建完整数据集 model_data = featured_data[available_features].copy() model_data = model_data.dropna()if len(model_data) == 0:print(f" 数据不足,跳过疾病 {disease}")continue# 划分训练集和测试集 train_data = model_data[model_data['year'] < 2016] test_data = model_data[model_data['year'] >= 2016]print(f" 训练集大小: {len(train_data)}, 测试集大小: {len(test_data)}")if len(train_data) == 0 or len(test_data) == 0:print(f" 数据不足,跳过疾病 {disease}")continue# 准备特征矩阵和目标向量 X_train = train_data.drop(columns=[disease]) y_train = train_data[disease] X_test = test_data.drop(columns=[disease]) y_test = test_data[disease]# 存储训练数据用于后续分析 X_train_list[disease] = X_train# 训练XGBoost模型print(" 训练XGBoost模型...")
# 预测 train_pred = xgb_model.predict(dtrain) test_pred = xgb_model.predict(dtest)# 计算评估指标 train_rmse = np.sqrt(mean_squared_error(y_train, train_pred)) test_rmse = np.sqrt(mean_squared_error(y_test, test_pred)) train_mae = mean_absolute_error(y_train, train_pred) test_mae = mean_absolute_error(y_test, test_pred) train_r2 = r2_score(y_train, train_pred) test_r2 = r2_score(y_test, test_pred)# 存储结果 disease_results = {'Disease': disease,'Train_Size': len(train_data),'Test_Size': len(test_data),'Train_RMSE': train_rmse,'Test_RMSE': test_rmse,'Train_MAE': train_mae,'Test_MAE': test_mae,'Train_R2': train_r2,'Test_R2': test_r2,'Best_Iteration': xgb_model.best_iteration } results_summary.append(disease_results)# 存储预测结果# 使用实际日期而不是索引 test_dates = featured_data.loc[ test_data.index, 'timestamp'] if'timestamp'in featured_data.columns else test_data.index pred_df = pd.DataFrame({'Date': test_dates,'Actual': y_test.values,'Predicted': test_pred,'Disease': disease }) predictions_list[disease] = pred_df# 存储模型和特征重要性 models_list[disease] = xgb_model# 获取特征重要性 importance_dict = xgb_model.get_score(importance_type='gain') importance_df = pd.DataFrame({'Feature': list(importance_dict.keys()),'Gain': list(importance_dict.values()) }).sort_values('Gain', ascending=False) feature_importance_list[disease] = importance_df# 可视化图表 try:
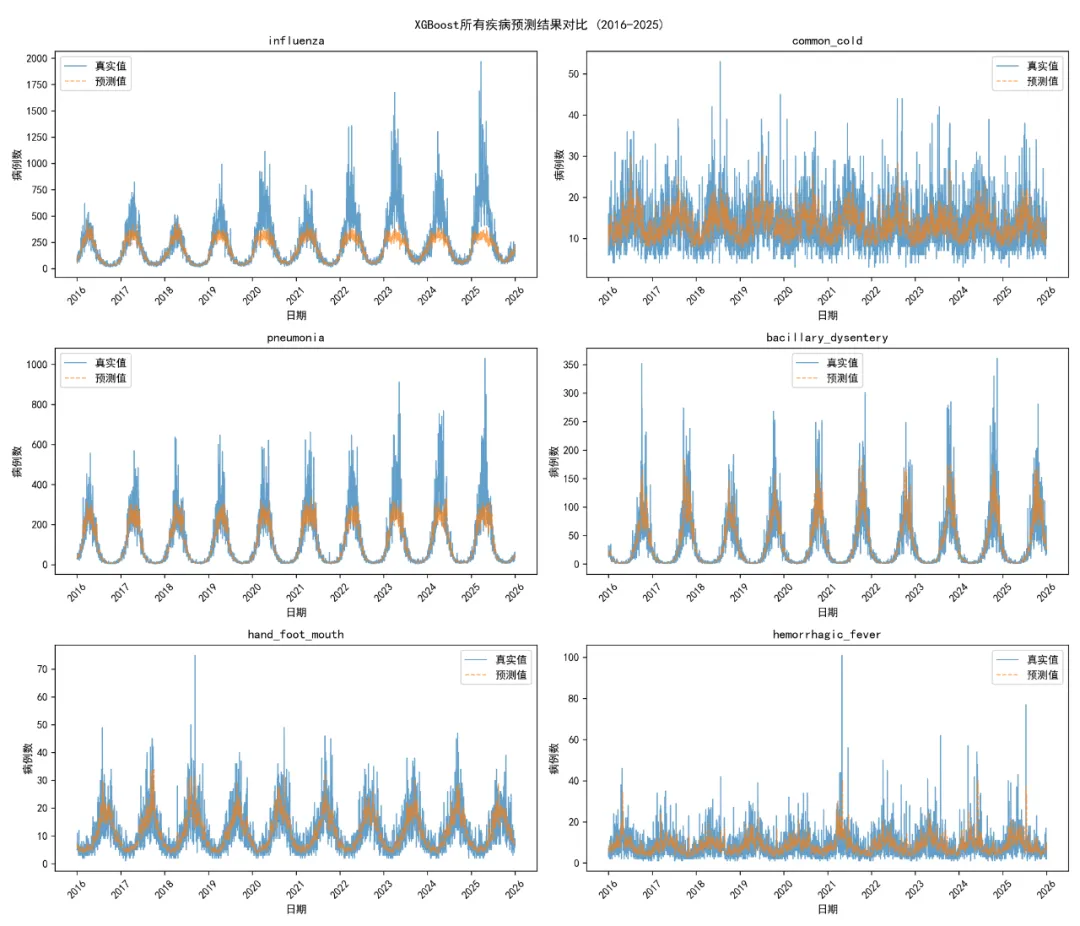
# 1. 预测结果对比图 plt.figure(figsize=(12, 6)) plt.plot(pred_df['Date'], pred_df['Actual'], label='真实值', linewidth=2, color='blue') plt.plot(pred_df['Date'], pred_df['Predicted'], label='预测值', linewidth=2, linestyle='--', color='red') plt.title(f'{disease} XGBoost预测结果对比\n测试集R² = {test_r2:.3f}, RMSE = {test_rmse:.1f}') plt.xlabel('日期') plt.ylabel('病例数') plt.legend() plt.xticks(rotation=45) plt.tight_layout() plt.savefig(os.path.join(results_path, f'Plots/{disease}_XGB_Results_Comparison.png'), dpi=300) plt.close()# 2. 特征重要性图 plt.figure(figsize=(10, 6)) top_features = importance_df.head(20) plt.barh(top_features['Feature'], top_features['Gain']) plt.title(f'{disease} XGBoost特征重要性 (前20)') plt.xlabel('Gain') plt.tight_layout() plt.savefig(os.path.join(results_path, f'Plots/{disease}_Feature_Importance.png'), dpi=300) plt.close()# 3. 实际值 vs 预测值散点图 plt.figure(figsize=(8, 6)) plt.scatter(pred_df['Actual'], pred_df['Predicted'], alpha=0.6, color='darkgreen') min_val = min(pred_df['Actual'].min(), pred_df['Predicted'].min()) max_val = max(pred_df['Actual'].max(), pred_df['Predicted'].max()) plt.plot([min_val, max_val], [min_val, max_val], 'r--', linewidth=2) plt.title(f'{disease} 实际值 vs 预测值') plt.xlabel('实际病例数') plt.ylabel('预测病例数') plt.tight_layout() plt.savefig(os.path.join(results_path, f'Plots/{disease}_Actual_vs_Predicted.png'), dpi=300) plt.close()# 4. 残差图 pred_df['Residuals'] = pred_df['Actual'] - pred_df['Predicted'] plt.figure(figsize=(8, 6)) plt.scatter(pred_df['Predicted'], pred_df['Residuals'], alpha=0.6, color='purple') plt.axhline(y=0, color='r', linestyle='--', linewidth=2) plt.title(f'{disease} 残差图') plt.xlabel('预测值') plt.ylabel('残差') plt.tight_layout() plt.savefig(os.path.join(results_path, f'Plots/{disease}_Residuals.png'), dpi=300) plt.close() except Exception as e:print(f" 可视化图表生成失败: {e}")# 保存特征重要性数据 importance_df.to_csv(os.path.join(results_path, f'Feature_Importance/{disease}_Feature_Importance.csv'), index=False)# SHAP分析print(" 生成SHAP分析图...") try: explainer = shap.TreeExplainer(xgb_model) shap_values = explainer.shap_values(X_train)# SHAP特征重要性图 plt.figure(figsize=(10, 8)) shap.summary_plot(shap_values, X_train, plot_type="bar", show=False) plt.title(f'{disease} SHAP特征重要性') plt.tight_layout() plt.savefig(os.path.join(results_path, f'SHAP_Analysis/{disease}_SHAP_Importance.png'), dpi=300) plt.close()# SHAP摘要图 plt.figure(figsize=(12, 8)) shap.summary_plot(shap_values, X_train, show=False) plt.title(f'{disease} SHAP值分布') plt.tight_layout() plt.savefig(os.path.join(results_path, f'SHAP_Analysis/{disease}_SHAP_Summary.png'), dpi=300) plt.close()# 保存SHAP值 shap_df = pd.DataFrame(shap_values, columns=X_train.columns) shap_df.to_csv(os.path.join(results_path, f'SHAP_Analysis/{disease}_SHAP_Values.csv'), index=False)# 计算SHAP重要性 shap_importance = pd.DataFrame({'Feature': X_train.columns,'Importance': np.mean(np.abs(shap_values), axis=0) }).sort_values('Importance', ascending=False) shap_importance.to_csv(os.path.join(results_path, f'SHAP_Analysis/{disease}_SHAP_Importance.csv'), index=False) except Exception as e:print(f" SHAP分析失败: {e}")print(f" {disease} 建模完成")# 保存总体结果if results_summary: results_df = pd.DataFrame(results_summary) results_df.to_csv(os.path.join(results_path, "Model_Performance_Summary.csv"), index=False)# 保存预测结果 all_predictions = pd.concat(predictions_list.values(), ignore_index=True) all_predictions.to_csv(os.path.join(results_path, "Predictions/All_Disease_Predictions.csv"), index=False)# 模型性能比较图 plt.figure(figsize=(12, 8)) results_sorted = results_df.sort_values('Test_R2')# 创建颜色映射 colors = plt.cm.RdYlGn((results_sorted['Test_R2'] - results_sorted['Test_R2'].min()) / (results_sorted['Test_R2'].max() - results_sorted['Test_R2'].min())) bars = plt.bar(range(len(results_sorted)), results_sorted['Test_R2'], color=colors, alpha=0.8) plt.title('XGBoost模型性能比较 (测试集)') plt.xlabel('疾病类型') plt.ylabel('R²') plt.xticks(range(len(results_sorted)), results_sorted['Disease'], rotation=45)# 添加数值标签for i, bar in enumerate(bars): height = bar.get_height() plt.text(bar.get_x() + bar.get_width() / 2., height + 0.01, f'{height:.3f}\nRMSE:{results_sorted.iloc[i]["Test_RMSE"]:.1f}', ha='center', va='bottom', fontsize=9) plt.tight_layout() plt.savefig(os.path.join(results_path, "Plots/Model_Performance_Comparison.png"), dpi=300) plt.close()# 所有疾病预测结果叠加图 fig, axes = plt.subplots(3, 2, figsize=(14, 12)) axes = axes.flatten()for i, (disease, pred_df) in enumerate(predictions_list.items()):if i < len(axes): ax = axes[i] ax.plot(pred_df['Date'], pred_df['Actual'], label='真实值', linewidth=1, alpha=0.7) ax.plot(pred_df['Date'], pred_df['Predicted'], label='预测值', linewidth=1, linestyle='--', alpha=0.7) ax.set_title(disease) ax.set_xlabel('日期') ax.set_ylabel('病例数') ax.legend() ax.tick_params(axis='x', rotation=45)# 隐藏多余的子图for i in range(len(predictions_list), len(axes)): axes[i].set_visible(False) plt.suptitle('XGBoost所有疾病预测结果对比 (2016-2025)') plt.tight_layout() plt.savefig(os.path.join(results_path, "Plots/All_Diseases_Predictions.png"), dpi=300) plt.close()
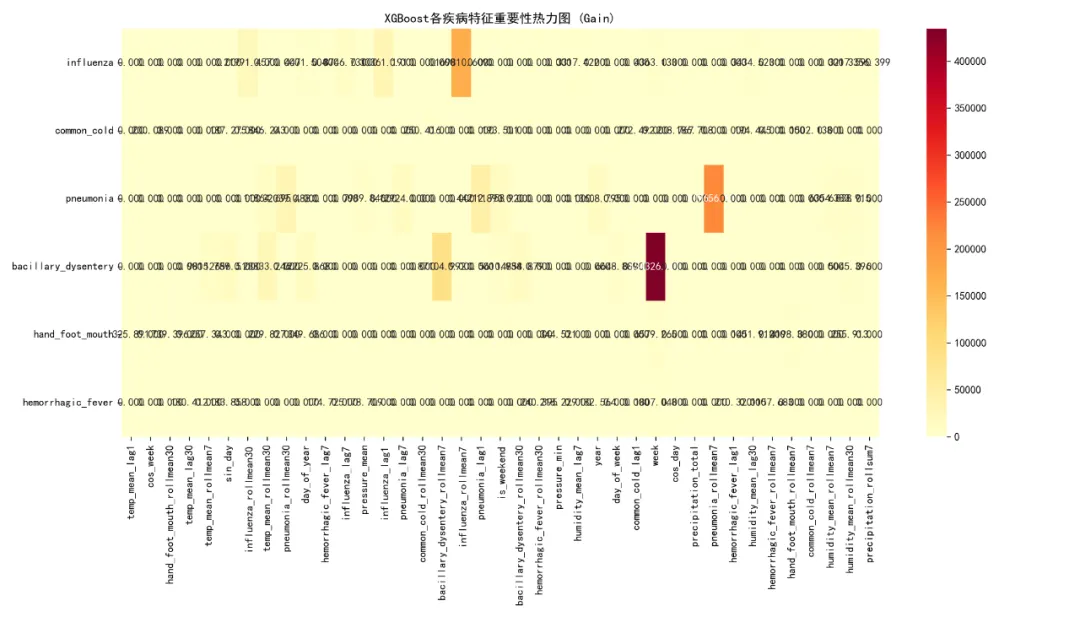
# 特征重要性热力图if len(feature_importance_list) > 0: try:# 收集所有特征 all_features = set()for imp_df in feature_importance_list.values(): all_features.update(imp_df['Feature'].head(10).tolist()) # 只取每个疾病的前10个特征# 创建重要性矩阵 importance_matrix = pd.DataFrame(0, index=available_diseases, columns=list(all_features))for disease, imp_df in feature_importance_list.items(): top_features = imp_df.head(10) # 只取前10个特征for _, row in top_features.iterrows():if row['Feature'] in all_features: importance_matrix.loc[disease, row['Feature']] = row['Gain']# 只保留有重要性的特征 non_zero_cols = importance_matrix.sum() > 0 importance_matrix = importance_matrix.loc[:, non_zero_cols]if len(importance_matrix.columns) > 0: plt.figure(figsize=(14, 8)) sns.heatmap(importance_matrix, annot=True, fmt='.3f', cmap='YlOrRd') plt.title('XGBoost各疾病特征重要性热力图 (Gain)') plt.tight_layout() plt.savefig(os.path.join(results_path, "Plots/Feature_Importance_Heatmap.png"), dpi=300) plt.close() except Exception as e:print(f"特征重要性热力图生成失败: {e}")# 保存模型for disease, model in models_list.items(): model.save_model(os.path.join(results_path, f'Models/{disease}_xgb.model'))# 生成最终报告print("\n=== XGBoost建模完成 ===")print(f"建模疾病数量: {len(models_list)}")print("训练集时间范围: 1981-2015")print("测试集时间范围: 2016-2025")print(f"总特征数量: {len(base_features) + len(available_diseases) * 5}\n")if results_summary: results_df = pd.DataFrame(results_summary)print("模型性能总结:")print(results_df.to_string(index=False)) best_model = results_df.loc[results_df['Test_R2'].idxmax()] worst_model = results_df.loc[results_df['Test_R2'].idxmin()]print(f"\n最佳性能模型:")print(f"疾病: {best_model['Disease']}, 测试集R²: {best_model['Test_R2']:.3f}, " f"RMSE: {best_model['Test_RMSE']:.1f}")print(f"\n最差性能模型:")print(f"疾病: {worst_model['Disease']}, 测试集R²: {worst_model['Test_R2']:.3f}, " f"RMSE: {worst_model['Test_RMSE']:.1f}")# 计算平均性能 avg_r2 = results_df['Test_R2'].mean() avg_rmse = results_df['Test_RMSE'].mean()print(f"\n平均性能:")print(f"平均测试集R²: {avg_r2:.3f}")print(f"平均测试集RMSE: {avg_rmse:.1f}")print("\n文件输出:")print(f"1. {results_path}/Model_Performance_Summary.csv - 模型性能汇总")print(f"2. {results_path}/Predictions/All_Disease_Predictions.csv - 所有预测结果")print(f"3. {results_path}/Models/ - 所有训练好的XGBoost模型")print(f"4. {results_path}/Plots/ - 各种可视化图表")print(f"5. {results_path}/Feature_Importance/ - 特征重要性数据")print(f"6. {results_path}/Partial_Dependence/ - 部分依赖图")print(f"7. {results_path}/SHAP_Analysis/ - SHAP分析结果")print(f"\n结果文件夹位置: {results_path}")if __name__ == "__main__": main()